Functions and iterations
2025-09-25
Spot a bug
df |> mutate(
a = (a - min(a, na.rm = TRUE)) /
(max(a, na.rm = TRUE) - min(a, na.rm = TRUE)),
b = (b - min(a, na.rm = TRUE)) /
(max(b, na.rm = TRUE) - min(b, na.rm = TRUE)),
c = (c - min(c, na.rm = TRUE)) /
(max(c, na.rm = TRUE) - min(c, na.rm = TRUE)),
d = (d - min(d, na.rm = TRUE)) /
(max(d, na.rm = TRUE) - min(d, na.rm = TRUE)),
)Functions improve resuability
normalize <- function(x) {
(x - min(x, na.rm = TRUE)) /
(max(x, na.rm = TRUE) - min(x, na.rm = TRUE))
}
df |> mutate(
a = normalize(a),
b = normalize(b),
c = normalize(c),
d = normalize(d),
)# A tibble: 5 × 4
a b c d
<dbl> <dbl> <dbl> <dbl>
1 0.564 0.161 0.208 0.391
2 0 0.215 0.253 0.723
3 0.231 0.327 1 1
4 0.299 1 0 0.524
5 1 0 0.222 0 Function syntax
Benefits of functions
Choose an evocative name that makes your code easier to understand.
As requirements change, you only need to update code in one place, instead of many.
You eliminate the chance of making incidental mistakes when you copy and paste (i.e. updating a variable name in one place, but not in another).
It makes it easier to reuse work from project-to-project, increasing your productivity over time.
Exercise I: improve normalize
Normalize based on user specified quantiles instead of min and max.
Exercise II: improve normalize
This new function breaks the existing code:
Error in `mutate()`:
ℹ In argument: `a = normalize(a)`.
Caused by error in `normalize()`:
! argument "probs" is missing, with no defaultHow to fix it?
Exercise II: improve normalize
We can add a default value so that legacy code (existing users of the function) doesn’t break.
normalize <- function(x, probs=c(0,1)) {
rng <- quantile(x, probs = probs, na.rm = TRUE)
(x - rng[1]) / (rng[2] - rng[1])
}
df |> mutate(
a = normalize(a),
b = normalize(b),
c = normalize(c),
d = normalize(d),
)# A tibble: 5 × 4
a b c d
<dbl> <dbl> <dbl> <dbl>
1 0.564 0.161 0.208 0.391
2 0 0.215 0.253 0.723
3 0.231 0.327 1 1
4 0.299 1 0 0.524
5 1 0 0.222 0 Higher-order functions
Functions that take other functions as arguments or return functions as their result.
base::apply function signature:
Xis an array (matrix)MARGINindicates whether the function will be applied over 1: rows or 2: columns.FUNis a function to be applied.
Function as an argument
…
... (dot-dot-dot) is a special argument, “catch-all”, that allows you to pass a variable number of arguments to a function.
[1] 5.5 5.0 4.5[1] 2.5 5.0 7.5na.rm is an argument to mean function, not apply.
optim the great
par: initial values for the parameters to be optimized.fn: the function to be minimized.gr: a function to compute the gradient offn. IfNULL, the gradient is approximated numerically....: arguments to be passed tofnandgr.
optim the great
m <- 1
s <- 3
log_gaussian_pdf <- function(x, mu, sd) {
-dnorm(x, mean = mu, sd = sd, log = TRUE)
}
results <- optim(0, log_gaussian_pdf, mu = m, sd = s, method="Brent", lower=-100, upper=100)
results$par[1] 0.9999999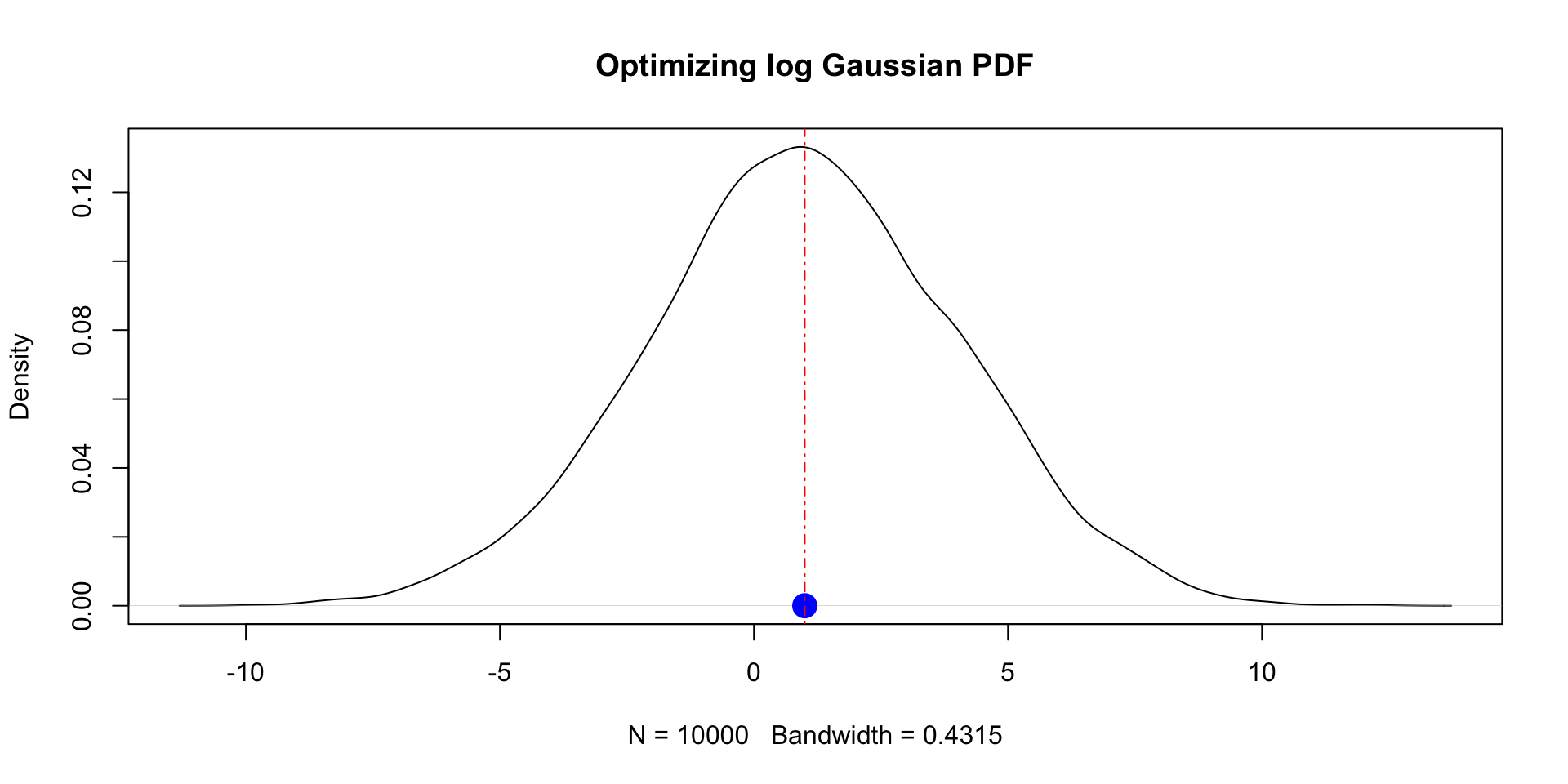
optim the great
Many uses:
- find maximum likelihood estimates or find posterior mode.
- minimize cost or loss functions (sum of square).
- can work with multivariate functions.
lambdas: anonymous functions
Some functions are needed only in a specific context and serve no further use.
If function x^2 + x + 3 is used once and never really again, we can define it inline without naming it.
Recursive functions
Function can call itself.
What does this function do?
Recursive functions
Components of a recursive function
Recursive functions are useful when for breaking down complex problem into simpler ones.
- Base Case: The condition that stops the recursion (e.g., if (n <= 1)). Without it, the function would call itself forever.
- Recursive Step: The part where the function calls itself (e.g., fib(n - 1) + fib(n - 2)).
- Recursion can be slow or even cause R to crash with a “stack overflow” error. This is because each function call uses a bit of memory, and too many nested calls can exhaust it.
Nested (or inner) function
What is the output of foo(2)?
Function factories
What is the output of f(3)?
Function indirection: get
What will be the output?
Function indirection: do.call
what: a function or a string naming the function to be called.args: a list of arguments to be passed to the function.
Function indirection: tidyverse
Programming with Tidyverse: use double curly braces { }.
set.seed(123)
df <- tribble(
~group, ~number,
"A", rnorm(1, mean = -5, sd = 1),
"A", rnorm(1, mean = -5, sd = 1),
"A", rnorm(1, mean = -5, sd = 1),
"B", rnorm(1, mean = 10, sd = 1),
"B", rnorm(1, mean = 10, sd = 1),
)
df# A tibble: 5 × 2
group number
<chr> <dbl>
1 A -5.56
2 A -5.23
3 A -3.44
4 B 10.1
5 B 10.1 Function indirection: tidyverse
compute_stat <- function(df, group_var, stat_var, func) {
df |> group_by({{ group_var }}) |> summarise(stat = func({{ stat_var }}, na.rm=TRUE))
}
compute_stat(df, group, number, mean)# A tibble: 2 × 2
group stat
<chr> <dbl>
1 A -4.74
2 B 10.1 # A tibble: 2 × 2
group stat
<chr> <dbl>
1 A -14.2
2 B 20.2Function scope
Variables defined inside a function are local to that function.
Function scope
increment_counter <- function(x) {
# 'counter' is created fresh every time the function is called
counter <- 0
counter <- counter + 1
return(x + counter)
}
increment_counter(10)[1] 11[1] 21No state persists across function calls.
Function scope: super assignment operator <<-
make_counter <- function() {
# 1. A variable is created in the parent environment
count <- 0
# 2. The factory returns a new, inner function
inner_function <- function() {
# 3. Use the super-assignment operator '<<-'
# This modifies 'count' in the parent environment, not locally.
count <<- count + 1#<<
return(count)
}
return(inner_function)
}Function scope: closure
- Inner function maintains the state even after parent function terminates.
Function scope: global environment
Iteration: for-loop
Iteration: while-loop
Iteration: repeat-loop
Vectorized operations
- Loops. R has to interpret each step of the loop one by one: check the condition, assign the value, increment the counter, and repeat. This involves a lot of overhead.
- Vectorized calls. R hands off the entire operation to highly optimized, pre-compiled code written in a lower-level language like C or Fortran. This code runs much closer to the machine level and executes the entire task in one efficient go.
Vectorized operations
num_samples <- 100000
time_loop <- system.time({
x <- numeric(num_samples)
for (n in 1:num_samples) {
x[n] <- rnorm(1) # Corrected from x[i] and specified rnorm(1)
}
})
time_vectorized <- system.time({
y <- rnorm(num_samples)
})
print(time_loop) # Loop time user system elapsed
0.209 0.047 0.262 user system elapsed
0.006 0.000 0.006 Testing with testthat
- Once you implement your function, you may want to verify that it works as expected.
- The main package for testing is
testthathttps://testthat.r-lib.org/. testthatoffers various convenient functions to check if your function behaves as expected.